Microbiologist Patrick McGann knew he had identified a dangerous germ. He just didn’t know how dangerous.
His job at the Walter Reed Army Institute of Research in Silver Spring, Md., is to prevent outbreaks of new pathogens in the U.S. military’s sprawling health-care system. In mid-May, a colleague found a strain of bacteria from a 49-year-old woman in Pennsylvania that had tested positive for resistance to a drug called colistin, the antibiotic of last resort.
McGann wasn’t yet alarmed. Sometimes mutations occur spontaneously that make bacteria resistant to an antibiotic. It would be a completely different problem, though, if the bacteria turned out to carry a certain colistin-resistance gene, called mcr-1, discovered in China late last year.
But that required deeper molecular testing. McGann asked his research team to do an initial test for the presence of the resistance gene. He was on Metro’s Red Line, headed back to the lab from a meeting, and wasn’t expecting the bombshell that followed.
On May 18 at 12:32 p.m., research technician Ana Ong texted him to say the strain of E. coli bacteria tested positive for the mcr-1 gene.
Stunned, McGann texted back: “You’re s——- me??” And then, realizing what the discovery meant, he didn’t wait for her reply.
“Onto the sequencer ASAP,” he wrote back, referring to the labor-intensive, round-the-clock effort that would have to come next to sequence the bacteria’s entire genome.
“We had to drop everything else to get this done,” he explained later. “When [the test] came back positive, all hell broke loose.”
Sequencing was the only way to answer the most critical question: How easily could the gene spread?
For infectious disease experts, the nightmare scenario is for the gene to spread to multidrug-resistant bacteria that cannot be killed by anything except colistin. That would make them invincible to any antibiotic, unstoppable by the most life-saving drugs of modern medicine.
—
The gene mcr-1 was found in China last November, first in pigs and then in people, alarming the infectious-disease community as it was reported across Asia and Europe. By March, there were more reports from France, Switzerland, Brazil and Argentina.
Still, the gene is relatively rare. After the initial report from China, McGann’s team tested more than 3,000 antibiotic-resistant bacteria that had been collected from military facilities around the world. None contained the mcr-1 gene.
The long-dreaded news that bacteria with the mcr-1 gene have been detected in the United States is giving new urgency to measures throughout the federal government to find where else the gene might be lurking and to control its spread.
The Centers for Disease Control and Prevention estimates that drug-resistant bacteria cause 2 million illnesses and about 23,000 deaths each year in the United States. Many soldiers wounded during the Iraq war contracted a strain of bacteria that the media later dubbed “Iraqibacter.” The normally harmless bacteria, found in soil and on skin, got into wounds and caused stubborn bloodstream infections, many of them resistant to many types of antibiotics. After about 2007, colistin was often one of the most effective treatment options available, McGann said.
That’s one reason why the Army created the Multidrug-resistant Organism Repository and Surveillance Network (MRSN) in 2009; McGann is its chief of molecular research and diagnostics. Researchers say the detection of colistin-resistant bacteria in the Pennsylvania woman was the result of this strong surveillance system but also sheer luck.
McGann got the initial call about a resistant E. coli sample from Kurt Schaecher, chief of the infectious diseases laboratory at Walter Reed National Military Medical Center in Bethesda. The Bethesda hospital has the reference lab for about 100 military facilities, which regularly send it drug-resistant organisms.
Schaecher, who had just started in the job, was troubled by the number of reports coming in from around the world about the mcr-1 gene. Testing for colistin resistance was not routinely done in the lab, but he thought it would be prudent to start.
On May 12, he pulled out six of the lab’s most recent samples to test for resistance to colistin. The next day, the results were back. “And lo and behold, this one popped up,” he recalled, referring to the Pennsylvania sample.
He immediately alerted McGann. Both men were very surprised, Schaecher said.
They didn’t immediately know why the bacteria weren’t killed by colistin – it could have been a spontaneous mutation rather than the mcr-1 gene. McGann’s team immediately went to Bethesda to collect the sample for more testing.
Once McGann’s lab detected the gene on May 18, he alerted Schaecher and began fully sequencing the bacteria’s genome.
—
Sequencing would tell everyone where the gene was located. Was it on the bacteria’s single chromosome? Or in a much more troubling spot, on something called a plasmid, a mobile piece of DNA that exists outside the chromosomes?
If the gene were on the chromosome, that wouldn’t be so bad. The bacteria could only pass the resistance to its progeny, limiting the spread. But finding the gene on the plasmid would be much, much worse. Plasmids can transfer their genes to other bacteria, McGann said, even strains that aren’t closely related. The technical term for this ability is “promiscuity.”
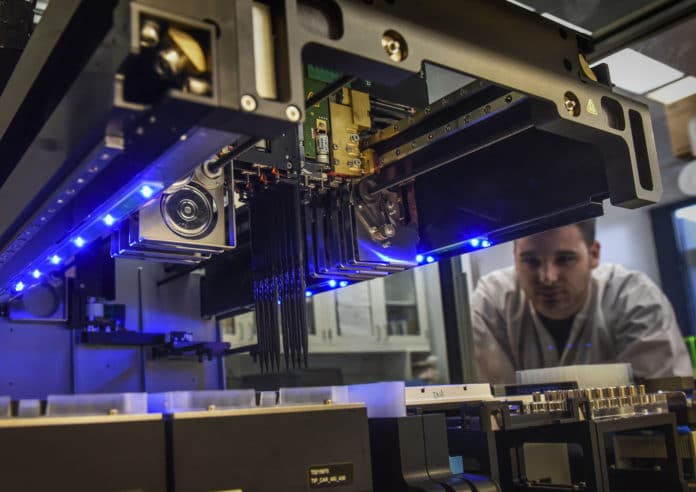
The sequencing process took several days. The researchers had to extract the DNA from the bacteria and set up the sequencing machines that would read the entire genome and determine not only what genes were in the organism, but also if there were plasmids and what kind. The researchers worked late, including Rosslyn Maybank, who is eight months pregnant.
By 9 p.m. on May 20, they were ready to proceed. They ran the same test on two machines. One sequencer is less accurate but takes about eight hours. The other larger sequencer, which costs about $700,000, is more accurate but takes 72 hours.
By the following day, early results confirmed the gene was fully present and was most likely on a plasmid. Barely 24 hours later, another researcher had confirmed that the mcr-1 was present “on a promiscuous plasmid,” McGann said. The final verification took until May 23.
“It was like finding a needle in a stack of needles in another stack of needles,” Schaecher recalled.
McGann likened plasmids to cars that can carry a gene and transport it to other bacteria. Some plasmids can transfer those genes more readily than others. “This one moved pretty well,” he said. “Not quite a Fiat, more like a BMW.”
The researchers began to notify colleagues in the Defense Department and other government agencies – the CDC and Health and Human Services and Agriculture departments – that the superbug had finally surfaced in a person in the United States and that it has the potential to spread colistin resistance far and wide.
Some officials have complained that they should have been informed sooner. McGann said it would have been imprudent to send an alert before results were completely confirmed. Around the same time, he, Schaecher and the research team submitted a paper about their findings to a medical journal. The editor, Yohei Doi, an infectious disease doctor at the University of Pittsburgh, raced to get it reviewed.
The paper was published May 26, and the discovery made headlines around the world.
—
Meanwhile, another surveillance system that includes the CDC, USDA and the Food and Drug Administration has also been searching for the gene in bacteria collected from food animals, meat sources and people. Scientists have scoured more than 44,000 samples of salmonella bacteria and 9,000 samples of E. coli and shigella bacteria.
That search is how USDA scientists recently found the gene in a sample from a pig intestine. It also was in a strain of E. coli, and also on a plasmid. USDA is working to determine the sample’s origin.
The strains and plasmids appear to be different, McGann said. That suggests that the gene is circulating through at least two – and possibly more – routes within the United States.
McGann said he only learned about the mcr-1 in the pig sample when his team notified government officials about its own finding. U.S. officials haven’t provided details about when the animal sample was found and why information about it wasn’t disclosed earlier.
Public health officials are most worried about the colistin-resistance gene spreading to a family of superbugs known as CRE, for carbapenem-resistant enterobacteriaceae, which the CDC has called one of the country’s most urgent public health threats. In some instances, CRE kills up to 50 percent of patients who become infected. Colistin is increasingly the last-resort drug to treat patients with such infections.
Already, doctors have been forced to rely on colistin as a last-line defense for many drug-resistant infections. Colistin, more than half a century old, is otherwise rarely used in human medicine because it can cause severe kidney damage.
The drug is not used in animals in the United States, but in Europe it has been widely used in veterinary medicine for decades to treat and prevent infection. In May, the European Medicines Agency recommended limiting its use in animals to halt the spread of resistance. It’s also widely used in farm animals in China.
The Army’s MRSN system receives between 400 to 500 samples of multidrug-resistant organisms each month, primarily from its bases around the world. About half are MRSA, or methicillin-resistant Staphylococcus aureus, a bacterium resistant to many antibiotics that can cause a range of problems, including skin infections and bloodstream infections. Another third are E. coli, and less than 1 percent are CRE.
The new information about the Pennsylvania woman’s colistin-resistant bacteria means McGann and his team can do more targeted searching in the repository of organisms. They’ve already scoured another 6,000 samples without again finding the gene.
Later this summer, all military services will be expected to begin providing the repository system with multidrug-resistant organisms.
By contrast, the national antibiotic resistance surveillance system that CDC, USDA and FDA operate collectively serves all civilian hospitals in the country. It covers a far larger patient population, and collects thousands of pathogen samples for each of the 15 antibiotic-resistant bacteria the CDC has deemed to be urgent or serious threats.
Until now, the resistance testing has taken place at CDC. But late last year, Congress approved $160 million in additional funding to boost the Obama administration’s antibiotic-resistance detection efforts. As a result, the agency plans to start funding state health departments this fall to do improved testing for antimicrobial resistance. That will include colistin.
—
Here’s how scientists discovered the antibiotic-resistant superbug:
http://wapo.st/28ffOSH